Children treated for B-Acute Lymphoblastic Leukemia (ALL) have high rates of recurrence. In order to overcome such challenges, patients require consolidation therapy that ranges from additional chemotherapy to hematopoietic stem cell transplant. Even so, approximately 50% of children with first relapse survive long term, which is important to determine the appropriate consolidation therapies. Currently, the most relevant risk stratification includes age, minimal residual disease (MRD) status seen in bone marrow 28 days post-induction, and cell count at the time of diagnosis. As the body of literature grows, there is a greater need to incorporate molecular data to assist in risk stratification.
In recent decades, microRNA (miRNA), which are non-coding RNA that regulate post-transcriptional modification, have been implicated in the pathogenesis of B-ALL. Due to the volume of variables, it is difficult to model such relationships using conventional methods. Thus, the objective was to generate a machine learning model to predict response to induction therapy by leveraging the Therapeutically Applicable Research to Generate Effect Treatment (TARGET) databases.
Between January 1, 2004 and January 1, 2011, the TARGET database collected data on over 11,000 patients with ALL up to the age of 31 that included clinical data such as age, gender, cytogenetics, and event-free survival (EFS). An event was defined as induction failure, relapse, progression, second hematologic malignancies, and death. In the expansion phase 2 part of the study, 175 children with B-ALL had in-depth genomic sequencing.
Because of the unpredictable downstream effects of dysregulation of miRNA, underlying assumptions of the Cox proportional hazard model may be violated. Machine learning models, such as the DeepSurv model, leverage deep neural networks to analyze survival data. The DeepSurv model has demonstrated superior performance in capturing linear and non-linear relationships between covariates and time-to-event analysis. In total, 171 features including age, MRD status, White Blood Cell (WBC) count at diagnosis, and 168 miRNA counts were included in the model. Random forest (RF) models were utilized to stratify the impact of features that are predictive.
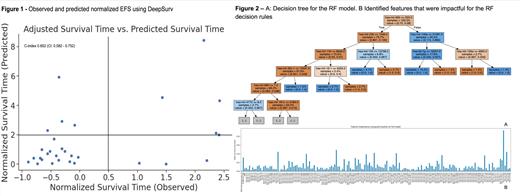
We created a 6773-parameter model using 171 features. A 10-fold cross-validation showed a concordance-index of 0.652 (CI: 0.582 - 0.752). Figure 1 shows the observed and predicted normalized EFS using DeepSurv. Notably, it is able to identify early relapse in the bottom L quadrant. Figure 2A shows the decision tree for the RF model that shows features and cut-offs as rules that cluster the EFS times. Figure 2B shows the features that were impactful for the RF decision rules.
In this work, we aim to leverage machine learning algorithms and miRNA to identify children who are at high risk for early relapse and can help clinicians to strategize consolidation therapy. We showed a reasonable concordance score that gives us a signal for potential impact. Further research, utilizing larger sample sizes, should serve to externally validate this model. Appropriate clinical application of such models has the potential to improve survival and limit treatment toxicity through a truly personalized approach to the patient and the unique genetic profile of their malignancy.
Disclosures
No relevant conflicts of interest to declare.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal